Current Projects
Organization of Genomic DNA
Genomic DNA in both prokaryotes and eukaryotes is compacted to fit into cellular compartments. We are interested in architectural proteins, so named because their primary function is to induce a specific DNA topology and control DNA compaction. Such proteins include the eukaryotic High Mobility Group (HMGB) proteins.
Architectural DNA-binding proteins play important roles in controlling processes such as DNA repair and gene expression. In eukaryotes, failure to regulate gene expression and DNA repair correctly may lead to mutagenesis, genomic instability, and cancer. Delineating these processes in bacteria is needed to understand how they respond to environmental changes, such as those associated with infection of a mammalian or plant host.
High Mobility Group proteins
 We are interested in the functional roles of High Mobility Group (HMGB) proteins in
yeast, Saccharomyces cerevisiae. HMGB proteins are generally known for their nuclear functions, altering chromatin
structure and regulating DNA-dependent activities. We are focusing on the HMGB homolog
Hmo1p, which resembles mammalian HMGB homologs by having two HMG-box domains. We have
shown that both HMG-boxes, box A and box B, of Hmo1p participate in DNA binding, and
that the lysine-rich C-terminal domain of Hmo1p is required for in-phase DNA bending
(Xiao et al., 2010).
We are interested in the functional roles of High Mobility Group (HMGB) proteins in
yeast, Saccharomyces cerevisiae. HMGB proteins are generally known for their nuclear functions, altering chromatin
structure and regulating DNA-dependent activities. We are focusing on the HMGB homolog
Hmo1p, which resembles mammalian HMGB homologs by having two HMG-box domains. We have
shown that both HMG-boxes, box A and box B, of Hmo1p participate in DNA binding, and
that the lysine-rich C-terminal domain of Hmo1p is required for in-phase DNA bending
(Xiao et al., 2010).
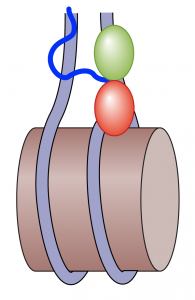 We have discovered that Hmo1p is important for stabilizing chromatin. Its deletion
creates a more dynamic chromatin environment in which remodeling events associated
with repair of DNA double strand breaks occur more rapidly (Panday et al., 2015). In this capacity, Hmo1p appears to function more like the mammalian linker histone
H1, which stabilizes chromatin in a process that requires its lysine-rich C-terminus;
while deletion of Hmo1p (or deletion of its basic C-terminal extension) renders chromatin
more sensitive to nuclease digestion, expression of human H1 in yeast deleted for
HMO1 restores chromatin resistance to nuclease, and rates of chromatin remodeling events
preceding DNA double strand break repair are also restored to wild-type levels (Panday & Grove, 2016).
We have discovered that Hmo1p is important for stabilizing chromatin. Its deletion
creates a more dynamic chromatin environment in which remodeling events associated
with repair of DNA double strand breaks occur more rapidly (Panday et al., 2015). In this capacity, Hmo1p appears to function more like the mammalian linker histone
H1, which stabilizes chromatin in a process that requires its lysine-rich C-terminus;
while deletion of Hmo1p (or deletion of its basic C-terminal extension) renders chromatin
more sensitive to nuclease digestion, expression of human H1 in yeast deleted for
HMO1 restores chromatin resistance to nuclease, and rates of chromatin remodeling events
preceding DNA double strand break repair are also restored to wild-type levels (Panday & Grove, 2016).
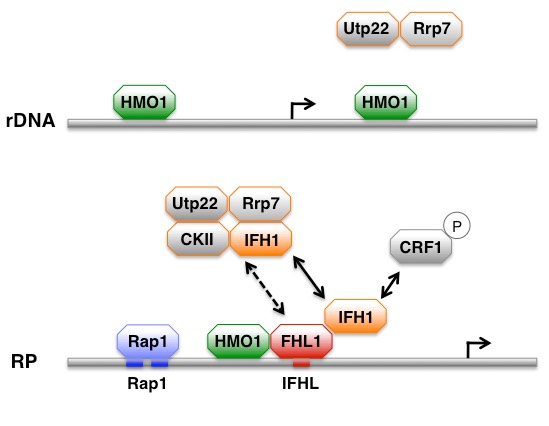
Current goals pertain to the mechanism by which Hmo1p participates in regulation of gene expression. Of specific interest is the role of Hmo1p in coordinating gene activity in response to signaling by the Target of Rapamycin (TOR) kinase pathway (Xiao et al., 2011). In keeping with Hmo1p being closely linked to mTORC1 activity, we discovered that TOR kinase binds directly to the HMO1 gene (Panday et al., 2017). For review see Panday & Grove, 2017; Grove, 2017; Kumar et al., 2022. While a mchanism by which mTORC1 regulates ribosomal protein genes has been suggested to depend on participation of the transcription factor Crf1p as a repressor, we have shown that Crf1p instead functions as an activator on specific ribosomal biogenesis genes, including HMO1 (Kumar et al., 2024).
Bacterial Responses to Stress
When a bacterial pathogen infects a host, a tug-of-war ensues in which the host aims to defend itself against the pathogen; the bacterium in turn responds to the new environmental cues, often subverting host defenses by utilizing host-derived signals to trigger upregulation of virulence genes. We are focusing on bacterial transcription factors that respond to such host-derived signals to control expression of virulence genes.
A major focus is on a group of transcriptional regulators designated MarR (Multiple Antibiotic Resistance Regulator) and the mechanism by which the binding of ligands controls their ability to regulate gene expression. Understanding mechanisms by which bacterial pathogens change gene expression programs in response to the environmental cues associated with host infection is critical for development of interventions required to combat bacterial infections.
MarR family transcriptional regulators
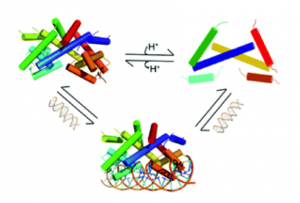 HucR. MarR proteins are transcriptional regulators that control gene activity in response
to stimuli such as ligand binding or cysteine oxidation (for review, see Deochand & Grove, 2017). We have characterized a MarR subfamily named UrtR for urate-responsive transcriptional
regulator. The founding member of this family is D. radiodurans HucR, which responds to urate by attenuated DNA binding in vitro and upregulation of uricase gene expression in vivo. The significance of this intricate control of uricase expression is that urate is
an efficient scavenger of ROS, thus maintaining optimal urate levels may contribute
to the oxidative stress resistance that is characteristic of D. radiodurans. We have also discovered that HucR is very pH-sensitive, forming a molten globule
state at pH~5 due to protonation of specific histidine residues at the dimer interface;
since DNA binding is abrogated at pH~5, the implication of this observation is the
potential for using HucR as a pH sensor in synthetic network devices (Deochand et al., 2016a).
HucR. MarR proteins are transcriptional regulators that control gene activity in response
to stimuli such as ligand binding or cysteine oxidation (for review, see Deochand & Grove, 2017). We have characterized a MarR subfamily named UrtR for urate-responsive transcriptional
regulator. The founding member of this family is D. radiodurans HucR, which responds to urate by attenuated DNA binding in vitro and upregulation of uricase gene expression in vivo. The significance of this intricate control of uricase expression is that urate is
an efficient scavenger of ROS, thus maintaining optimal urate levels may contribute
to the oxidative stress resistance that is characteristic of D. radiodurans. We have also discovered that HucR is very pH-sensitive, forming a molten globule
state at pH~5 due to protonation of specific histidine residues at the dimer interface;
since DNA binding is abrogated at pH~5, the implication of this observation is the
potential for using HucR as a pH sensor in synthetic network devices (Deochand et al., 2016a).
PecS. We have identified the residues involved in coordinating urate in HucR (Perera et al., 2009). Notably, a group of MarR homologs conserve these residues; this subfamily includes PecS proteins, transcription factors involved in regulating expression of virulence genes in certain plant pathogens (Perera & Grove, 2011). We have shown that repression of gene expression by PecS from Agrobacterium fabrum, which causes crown gall tumors in a variety of host plants, Pectobacterium atrosepticum, which infects potato, the soil bacterium Streptomyces coelicolor, and Vibrio vulnificus, which is best known for causing gastrointestinal distress after eating raw oysters, is relieved in presence of urate and/or xanthine (Perera & Grove, 2010; Huang et al., 2013; Deochand et al., 2016b; Bhattacharyya et al., 2019; Nwokocha et al., 2023). In A. fabrum, urate levels remain low even under stress conditions, suggesting that it is urate produced by the host that functions as an inducing ligand (Sivapragasam et al., 2017).
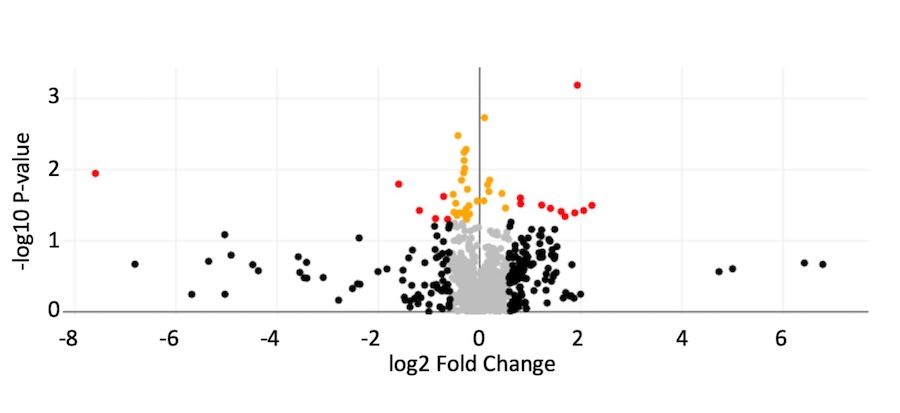 PecS regulates expression of pecM, which encodes an efflux pump. We reported that A. fabrum PecM exports the toxic plant exudate, 4-hydroxybenzaldehyde. This was established
in part by using untargeted metabolomics to compare wild-type and pecM disruption strains, an experiment that showed accumulation of 4-hydroxybenzaldehyde
as well as certain intermediates in the citric acid cycle when PecM is absent. These
data indicate that PecM is important for maintaining cellular homeostasis (Ghosh & Grove, 2025). By comparison, disruption of A. fabrum PecS revealed that PecS is important for controlling features such as survival in
microaerobic conditions and accumulation of acyl homoserine lactone (Nwokocha et al., 2023). Taken togeter, our data indicate that A. fabrum PecS and PecM both promote bacterial fitness in the rhizosphere. For a recent review
about PecS proteins, see Nwokocha et al., 2024. A current focus is to define the A. fabrum PecS regulon.
PecS regulates expression of pecM, which encodes an efflux pump. We reported that A. fabrum PecM exports the toxic plant exudate, 4-hydroxybenzaldehyde. This was established
in part by using untargeted metabolomics to compare wild-type and pecM disruption strains, an experiment that showed accumulation of 4-hydroxybenzaldehyde
as well as certain intermediates in the citric acid cycle when PecM is absent. These
data indicate that PecM is important for maintaining cellular homeostasis (Ghosh & Grove, 2025). By comparison, disruption of A. fabrum PecS revealed that PecS is important for controlling features such as survival in
microaerobic conditions and accumulation of acyl homoserine lactone (Nwokocha et al., 2023). Taken togeter, our data indicate that A. fabrum PecS and PecM both promote bacterial fitness in the rhizosphere. For a recent review
about PecS proteins, see Nwokocha et al., 2024. A current focus is to define the A. fabrum PecS regulon.
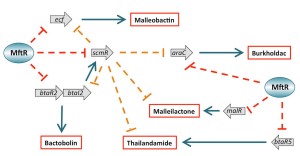 MftR. MftR proteins constitute another group of urate-responsive transcription factors.
MftR is for example encoded by Burkholderia species, some of which are serious human pathogens. Based on analysis of MftR, we
have reported that residues that are invariable among UrtR homologs are conserved
for structural reasons (Gupta & Grove, 2014). We have used RNA-seq to identify the MftR regulon in B. thailandensis. Notably, MftR acts as a global repressor of biosynthetic gene clusters encoding
proteins responsible for synthesis of secondary metabolites, many of which improve
bacterial fitness and have pharmaceutical importance. Since such gene clusters are
typically silenced under laboratory conditions, this observation paves the way for
identification of novel therapeutic leads (Gupta et al., 2017; Thapa & Grove, 2019; Thapa et al., 2022). Notably, the antibiotic trimethoprim elicits the accumulation of xanthine, which
is also a ligand for MftR. This in turn leads to de-repression of genes under MftR
control, a consequence of which is increased virulence (Thapa & Grove, 2020). We are currently working on identifying direct MftR targets.
MftR. MftR proteins constitute another group of urate-responsive transcription factors.
MftR is for example encoded by Burkholderia species, some of which are serious human pathogens. Based on analysis of MftR, we
have reported that residues that are invariable among UrtR homologs are conserved
for structural reasons (Gupta & Grove, 2014). We have used RNA-seq to identify the MftR regulon in B. thailandensis. Notably, MftR acts as a global repressor of biosynthetic gene clusters encoding
proteins responsible for synthesis of secondary metabolites, many of which improve
bacterial fitness and have pharmaceutical importance. Since such gene clusters are
typically silenced under laboratory conditions, this observation paves the way for
identification of novel therapeutic leads (Gupta et al., 2017; Thapa & Grove, 2019; Thapa et al., 2022). Notably, the antibiotic trimethoprim elicits the accumulation of xanthine, which
is also a ligand for MftR. This in turn leads to de-repression of genes under MftR
control, a consequence of which is increased virulence (Thapa & Grove, 2020). We are currently working on identifying direct MftR targets.
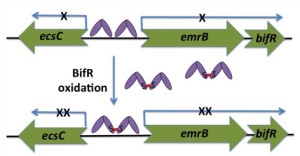 Responses to other host signals. Production of ROS is a common host defense that is elicited upon pathogen infection.
By contrast, for bacteria that establish biofilm communities, oxygen may become limiting.
Another active area of research focuses on MarR homologs that respond to changes in
(reactive) oxygen levels to alter gene expression programs upon host infection. One
example is B. thailandensis BifR, which is converted to a dimer-of-dimers "super-repressor" upon oxidation and
controls biofilm formation (Gupta et al., 2017). For a review on MarR proteins from Burkholderia species, see Gupta et al., 2019.
Responses to other host signals. Production of ROS is a common host defense that is elicited upon pathogen infection.
By contrast, for bacteria that establish biofilm communities, oxygen may become limiting.
Another active area of research focuses on MarR homologs that respond to changes in
(reactive) oxygen levels to alter gene expression programs upon host infection. One
example is B. thailandensis BifR, which is converted to a dimer-of-dimers "super-repressor" upon oxidation and
controls biofilm formation (Gupta et al., 2017). For a review on MarR proteins from Burkholderia species, see Gupta et al., 2019.
Using quantitative mass spectrometry to determine proteome changes imposed in strains deleted for a specific transcription factor
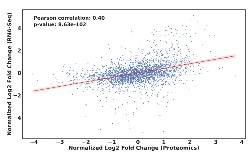 A current focus is on defining differentially expressed proteins as a result of deleting
a specific transcription fator. While RNA sequencing was instructive in terms of defining
the B. thailandensis MftR regulon, differential accumulation of mRNAs does not equal differential protein
expression. For instance, we recently reported that when B. thailandensis enters stationary phase, a large number of both mRNAs and proteins differentially
accumulate. However, an only 40% correlation between changes in mRNA and protein levels
were seen, which speaks to marked post-transcriptional regulation (Al-Tohamy et al.,
2025). We are therefore implementing quantitative mass spectrometry for a more incisive
determination of transcription factor function.
A current focus is on defining differentially expressed proteins as a result of deleting
a specific transcription fator. While RNA sequencing was instructive in terms of defining
the B. thailandensis MftR regulon, differential accumulation of mRNAs does not equal differential protein
expression. For instance, we recently reported that when B. thailandensis enters stationary phase, a large number of both mRNAs and proteins differentially
accumulate. However, an only 40% correlation between changes in mRNA and protein levels
were seen, which speaks to marked post-transcriptional regulation (Al-Tohamy et al.,
2025). We are therefore implementing quantitative mass spectrometry for a more incisive
determination of transcription factor function.