Research
The LSU AgCenter Small Grains program focuses on variety development and quantitative genetics methods that support and accelerate the variety development pipeline. Primary efforts investigate optimal strategies for using molecular markers to improve breeding for pest and pathogen resistance using both marker assisted selection and genomic prediciton. While genetic resistance to these biotic stresses are generally repeatable from year to year, variation for grain yield is affected by those biotic stresses and all other events that occur in a plot's life history, inclulding abiotic stresses such as heat, flood, and drought that are impacted by the specific environmental conditions of a location and year. Modern database tools are being used to gather together far-ranging historic data sets on phenotypes, genotypes, and environmental variables to create data sets useful for investigating these challenges. Better understanding the yield effects of major genes controlling wheat phenology, that through phenological variation generate environment-specific yield variation, is a major focus of this work. Beyond major genes, machine learning approaches that integrate interactions between environmental variables and genetic data allow for the prediction of site-specific yield, and have become a heavily-used tool within the LSU AgCenter wheat breeding program. Work continues on optimizing those models and the data that powers them by studying optimal early-generation, multi-location trial design.
Research Areas
Marker-based breeding for pest and pathogen resistance

- Marker development, fine-mapping, and validation of major Hessian fly resistance genes and development of adapted wheat lines carrying Hessian fly gene pyramids.
- Genomic prediction and sparse testing approaches to improve quantitative field resistance of wheat to Hessian fly.
- Optimizing genomic prediction models for Fusarium head blight resistance, including incorporation of information on known major resistance QTL.
- Identification of novel resistance genes for oat crown rust and oat stem rust in wild oat relatives.
Wheat abiotic stress and phenology
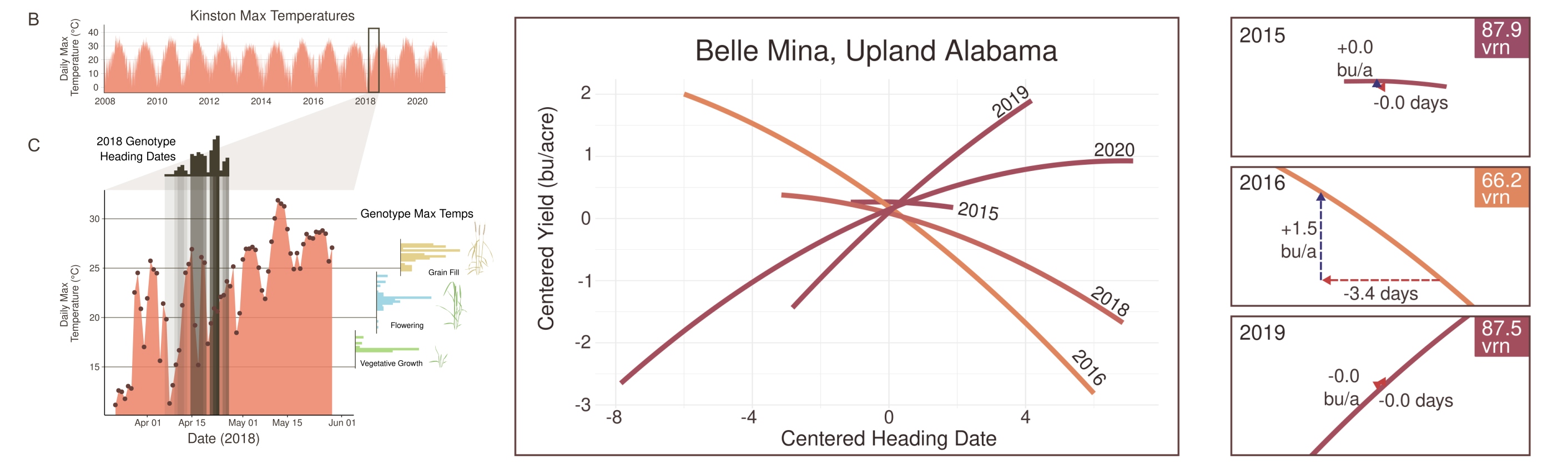
- Optimizing wheat phenology for double-cropping of wheat with soybeans, understanding the impact of varying wheat maturities with soybean planting date and joint profitability of the double-crop system.
- Relationship of major wheat phenology genes (Vrn and Ppd) with environment-specific yield effects as mediated by environmental variables such as heat, freeze, and drought stress.
Optimizing genomic selection and field testing
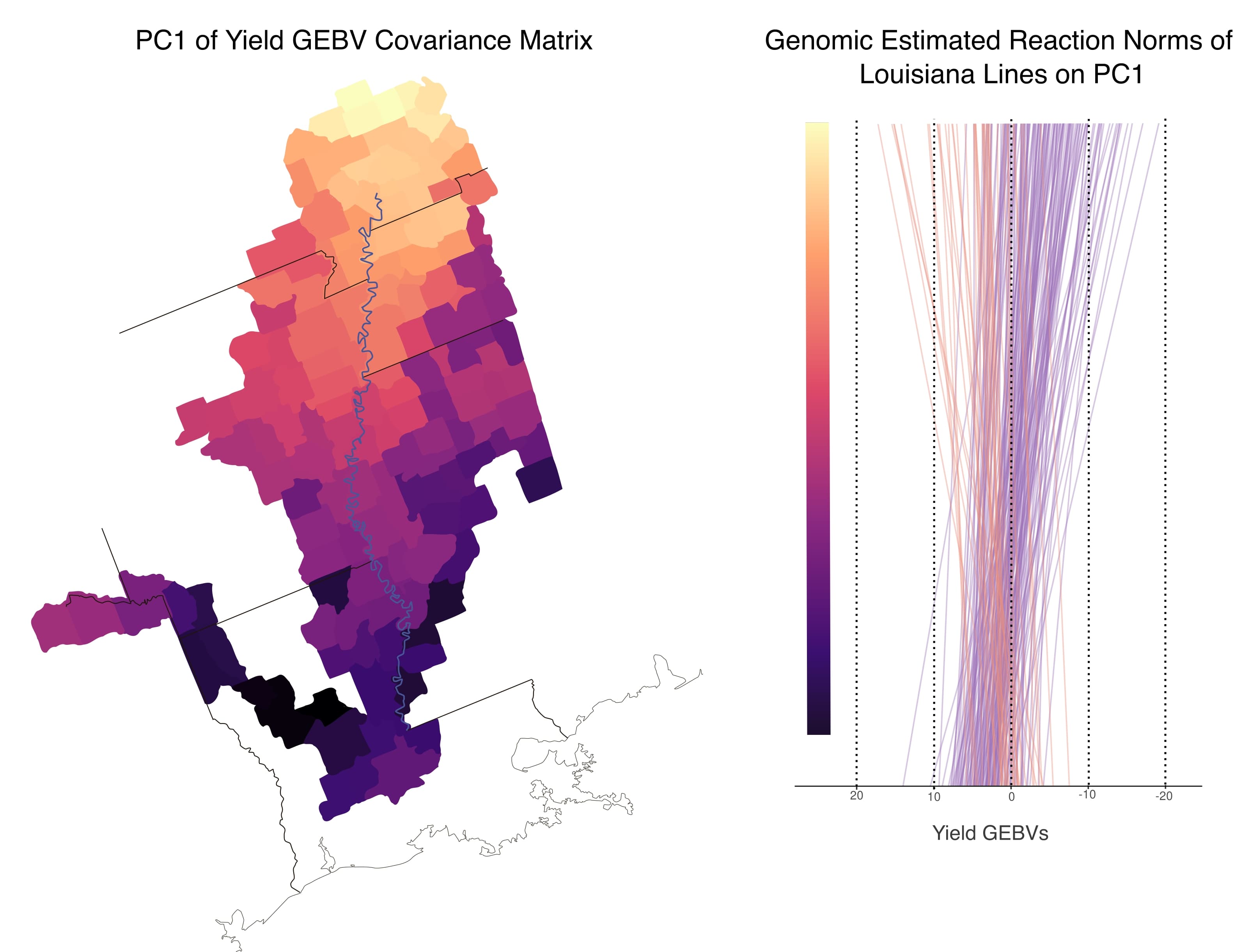
- Incorporating environmental variables into genomic predictions from historic data for wheat yield to target specific macroenvironments within the broader Southern United States.
- Genomics-enabled sparse testing designs for multi-region breeding programs.
- Optimizing within-family evaluation of early-generation breeding program lines for Fusarium head blight resistance.
Digital infrastructure for breeding programs
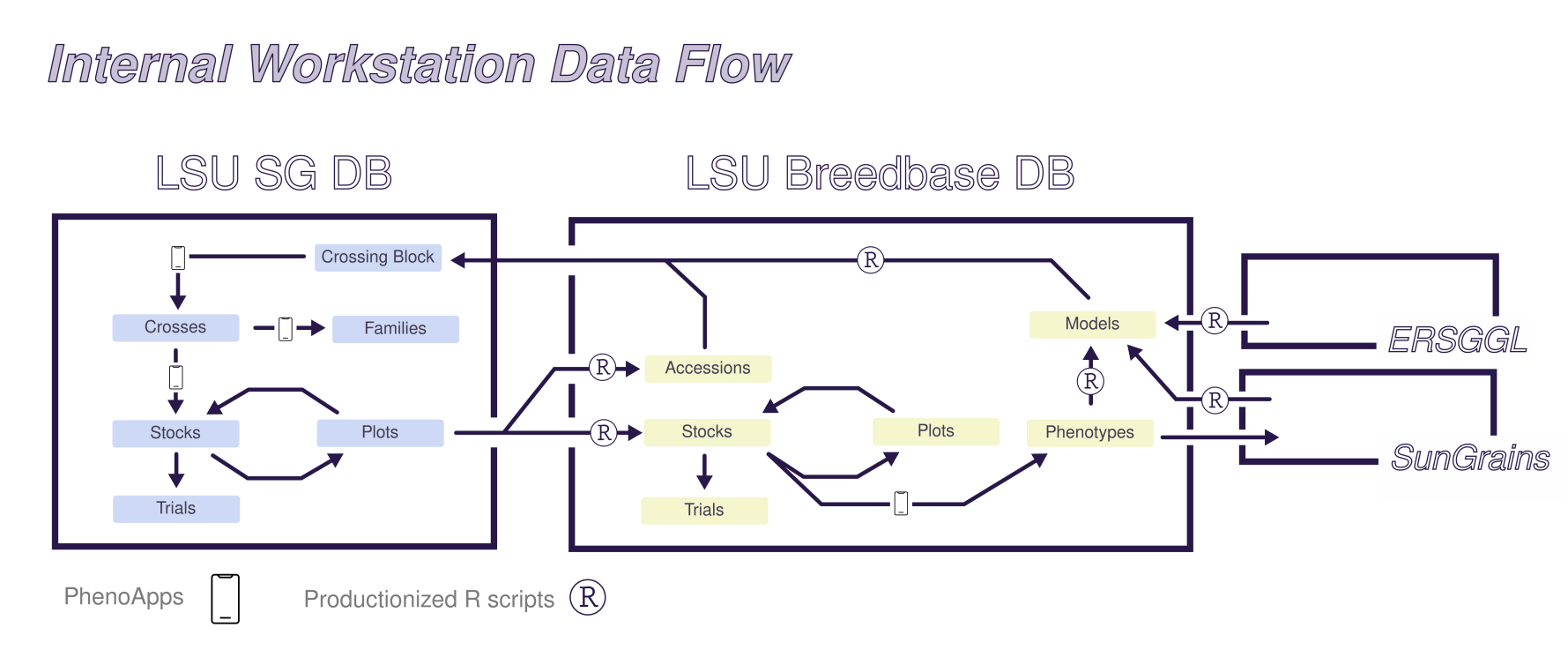
- BrAPI-based tools for interfacing with Breedbase for applied breeding programs.
- Database software for line-development portion of pedigreed breeding programs.
- Recurring updates of family-based genomic predictions based on pedigree information.
Cooperative and Variety Testing Reports
- 2024 Wheat variety trial report
- 2024 Oat variety trial report
- 2024 Uniform Southern Soft Red Winter Wheat Report
- 2024 Uniform Southern Scab Report
Variety Releases
- Md Ali Babar*, Naeem Khan, Ann Blount, Ronald D Barnett, Stephen A Harrison, Noah DeWitt, Jerry Johnson, Mohamed Mergoum, Rick Boyles, Paul Murphy, Esten Mason, Ehsan Shakiba, Amir MH Ibrahim, Russell Sutton, Gina Brown‐Guedira, David Marshall, Christina Cowger, Byung‐Kee Baik, Nicholas Santantonio, Sue Cambron, Yue Jin, Daniel Mailhot. Registration of ‘FL16045‐25’: An early‐maturing, high‐yielding, disease‐resistant soft red facultative wheat cultivar for the southern United States (2024). Journal of Plant Registrations
- Md Ali Babar*, Stephen A Harrison, Ann Blount, Ronald D Barnett, Jerry Johnson, Mohamed Mergoum, Daniel J Mailhot, J Paul Murphy, Richard E Mason, Ehsan Shakiba, Amir MH Ibrahim, Russell Sutton, Bryan Simoneaux, Richard Boyles, Noah DeWitt, Brad Stancil, David Marshall, Myron Fountain, Kathy Esvelt Klos, Naeem Khan, Marcelo Wallau, Henry G Jordan Jr, Xuefeng Ma, Juan Arbelaez. Registration of ‘FL12034‐10’oat: A new dual‐purpose disease resistant cultivar for Florida and southern United States (2024). Journal of Plant Registrations
Publications
- Noah DeWitt*, Jeanette Lyerly, Mohammed Guedira, James B Holland, J Paul Murphy, Brian P Ward, Richard E Boyles, Mohamed Mergoum, Md Ali Babar, Ehsan Shakiba, Russel Sutton, Amir Ibrahim, Vijay Tiwari, Nicholas Santantonio, David A Van Sanford, Kimberly Howell, Jared H Smith, Stephen A Harrison, Gina Brown-Guedira. Bearded or Smooth? Awns Improve Yield when Wheat Experiences Heat Stress During Grain Fill in the Southeastern United States (2023). Journal of Experimental Botany.
- Aifeng Liu, Jill A. Nemacheck, Hongwei Li, Kirk M. Anderson, Noah DeWitt, Marion O. Harris, Steven S. Xu, Subhashree Subramanyam*. Triticum monococcum subsp. monococcum and aegilopoides: new sources of resistance to the dipteran pest, Hessian fly (Diptera: Cecidomyiidae) (2024). Journal of Economic Entomology.
- Zachary J. Winn*, Amanda L. Amsberry, Scott D. Haley, Noah DeWitt, R. Esten Mason. Phenomic Versus Genomic Prediction - A Comparison of Prediction Accuracies for Grain Yield in Hard Winter Wheat Lines (2023). The Plant Phenome Journal.
- Zachary J. Winn*, Emily Hudson‐Arns, Mikayla Hammers, Noah DeWitt, Jeanette Lyerly, Guihua Bai, Paul St Amand, Punya Nachappa, Scott Haley, Richard Esten Mason. HaploCatcher: An R package for prediction of haplotypes (2024). The Plant Genome
- Richard Boyles*, Carolina Ballén‐Taborda, Gina Brown‐Guedira, Jose Costa, Christina Cowger, Noah DeWitt, Carl Griffey, Steven Harrison, Amir Ibrahim, Jerry Johnson, Jeanette Lyerly, David S. Marshall, R. Esten Mason, Mohamed Mergoum, J. Paul Murphy, Nicholas Santantonio, Gautam Saripalli, Russell Sutton, Vijay Tiwari, David van Sanford, Zachary J. Winn. Approaching 25 years of progress towards Fusarium head blight resistance in southern soft red winter wheat (Triticum aestivum L.) (2023). Plant Breeding.
- Noah DeWitt*, Mohammed Guedira, J. Paul Murphy, David Marshall, Mohamed Mergoum, Christian Maltecca, Gina Brown-Guedira. A network modeling approach provides insights into the environment-specific yield architecture of wheat (2022). Genetics 221 (3).
- Zachary J. Winn*, Dylan L. Larkin, Dennis N. Lozada, Noah DeWitt, Gina Brown-Guedira, Richard Esten Mason. Multivariate genomic selection models improve prediction accuarcy of agronomic traits in soft red winter wheat (2023). Crop Science 63 (4), 2115-2130.
- Fei He, Wei Wang, William B Rutter, Katherine W Jordan, Jie Ren, Ellie Taagen, Noah DeWitt, Deepmala Sehgal, Sivakumar Sukumaran, Susanne Dreisigacker, Matthew Reynolds, Jyotirmoy Halder, Sunish Kumar Sehgal, Shuyu Liu, Jianli Chen, Allan Fritz, Jason Cook, Gina Brown-Guedira, Mike Pumphrey, Arron Carter, Mark Sorrells, Jorge Dubcovsky, Matthew J Hayden, Alina Akhunova, Peter L Morrell, Les Szabo, Matthew Rouse, Eduard Akhunov*. Genomic variants affecting homoeologous gene expression dosage contribute to agronomic trait variation in allopolyploid wheat (2022). Nature Communications 13 (1), 1-15.
- Noah DeWitt*, Mohammed Guedira, Edwin Lauer, J Paul Murphy, David Marshall, Mohamed Mergoum, Jerry Johnson, James B Holland, Gina Brown-Guedira. Characterizing the oligogenic architecture of plant growth phenotypes informs genomic selection approaches in a common wheat population (2021). BMC genomics 22 (1), 1-18.
- Noah DeWitt, Mohammed Guedira, Edwin Lauer, Martin Sarinelli, Priyanka Tyagi, Daolin Fu, QunQun Hao, J. Paul Murphy, David Marshall, Alina Akhunova, Katherine Jordan, Eduard Akhunov, Gina Brown-Guedira*. Sequence‐based mapping identifies a candidate transcription repressor underlying awn suppression at the B1 locus in wheat (2019). New Phytologist 225.1: 326-339.