Research & Publications
Research
The common theme of our research is investigating the structure-function relationship of proteins as it relates to activity, regulation, and signaling. Our primary tool for high-resolution, three-dimensional structure determination is nuclear magnetic resonance (NMR) spectroscopy. We also use mass spectrometry (MS), surface plasmon resonance (SPR), x-ray crystallography, fluorescence, and chromatography methods.
|
|
|
|
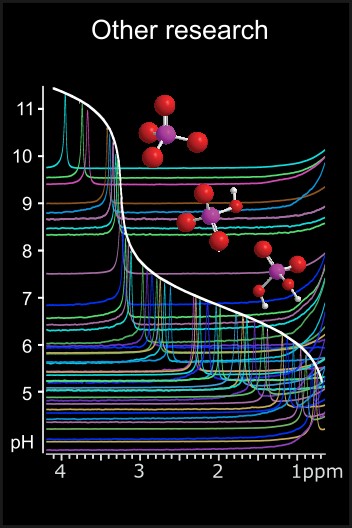
Other Research Interests
|
|
|
|
|
|
|
|
Publications
(* corresponding author, † undergraduate author)
-
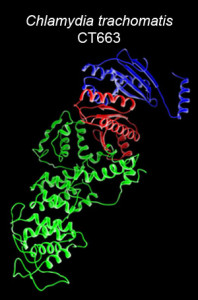
T. O. Ukwaththage, S. Keane,† Li Shen, and M. A. Macnaughtan,* Chain-selective istopic labeling of the heterodimeric type III secretion chaperone, Scc4:Scc1, reveals the total structural rearrangement of the Chlamydia trachomatis bi-functional protein, Scc4. Biomolecules, 2020, 10.
-
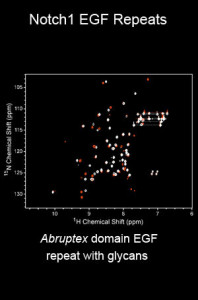
J. A. Grennell, K. D. Jenkins, H. Zhong, A. Paudyal, K. B. Luther, R. S. Haltiwanger, M. A. Macnaughtan,* Expression, purification, and glycosylation of epidermal growth factor-like repeat 27 from mouse NOTCH1. Protein Expression and Purification, 2020, 174, 105681-105692.
-
T. O. Ukwaththage, T. O. Tonelli, and M. A. Macnaughtan,* Backbone and sidechain resonance assignments and secondary structure of Scc4 from Chlamydia trachomatis. Biomolecular NMR Assignments, 2020, 14, 301-307.
-
L. Gao, Y. Cong, G. V. Plano, X. Rao, L. Gisclair, S. Schesser Bartra, M. A. Macnaughtan,* and L. Shen,* Context-dependent action of Scc4 reinforces control of the type III secretion system. Journal of Bacteriology, 2020, 202(115), e00132-20.
-
T. O. Ukwaththage, O. Y. Goodwin, A. C. Songok, A. M. Tafaro,† L. Shen, and M. A. Macnaughtan*, Purification of tag-free Chlamydia trachomatis Scc4 for structural studies using sarkosyl-assisted on-column complex dissociation. Biochemistry, 2019, 58(42), 4284-4292.
-
A. C. Songok, P. Panta, W. T. Doerrler, M. A. Macnaughtan, and C. M. Taylor, Structural modification of the tripeptide KPV by reductive “glycoalkylation” of the lysine residue. PLoS One, 2018, 13(6), 1-14.
-
C. E. Lane, O. Y. Goodwin, M. A. Macnaughtan, and M. G. Benton, Novel interpretations of in vitro polyhydroxyalkanoate polymerization phenomena. Polymer, 2016, 103, 196-205.
-
L. Shen,* M. A. Macnaughtan, K. M. Frohlich, Y. Cong, O. Y. Goodwin, C.-W. Chou, L. LeCour, K. L. Krup, M. Luo, D. K. Worthylake, Multipart chaperone-effector recognition in the type III secretion system of Chlamydia trachomatis. Journal of Biological Chemistry, 2015, 290, 28141-28155.
-
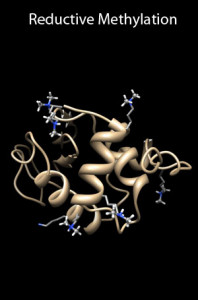
P. N. Brady and M. A. Macnaughtan,* Evaluation of colorimetric assays for analyzing reductively methylated proteins: Biases and mechanistic insights. Analytical Biochemistry, 2015, 491, 43-51.
-
H. Negulescu, Y. Guo, T. P. Garner, O. Y. Goodwin, G. Henderson, R. A. Laine, M. A. Macnaughtan,* A Kazal-type serine protease inhibitor from the defense gland secretion of the subterranean termite Coptotermes formosanus Shiraki. PLoS ONE, 2015, 10(5):e0125376.
-
H. Negulescu, Y. Guo, T. P. Garner, O. Y. Goodwin, G. P. Henderson, R. A Laine, and M. A. Macnaughtan,* NMR structure of a Kazal-type serine protease inhibitor from the subterranean termite defense gland of Coptotermes formosanus Shiraki soldiers. Protein Data Bank, 2015, PDB ID: 2N17 (formerly 2M25).
-
E. B. Fontenot, S. F. Ditusa, N. Kato, D. M. Olivier, R. Dale, W. Y. Lin, T. J. Chiou, M. A. Macnaughtan, and A. P. Smith,* Increased phosphate transport of Arabidopsis thaliana Pht1;1 by site-directed mutagenesis of tyrosine 312 may be attributed to the disruption of homomeric interactions. Plant, Cell & Environment, 2015, 38, 2012-2022.
-
K. J. Roberson and M. A. Macnaughtan,* Review: Methods to assign NMR resonances from reductively methylated proteins. Analytical Biochemistry, 2014, 466, 76-82.
-
S. Ghezal, M. S. Thomasson, I. Lefebvre-Tournier, C. Périgaud, M. A. Macnaughtan, and B. Roy,* CDP-ethanolamine and CDP-choline: One-pot synthesis and 31P NMR study. Tetrahedron Letters, 2014, 55(38), 5306-5310.
-
M. S. Thomasson and M. A. Macnaughtan,* Review: Microscopy basics for the study of actin – actin-binding protein interactions, Analytical Biochemistry, 2013, 443(12), 156-165.
-
K. J. Roberson and M. A. Macnaughtan,* Attempts towards unambiguously assigning 13C-dimethylamine NMR resonances. The All Results Journal: Chemistry, 2013, 4, 10-16.
-
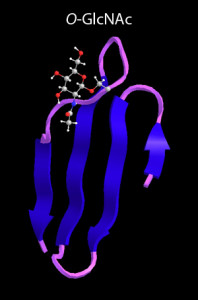
O. Y. Goodwin, M. M. Sweeney, M. S. Thomasson, A. J. Lin,† and M. A. Macnaughtan,* E. coli sabotages the in vivo production of O-linked beta-N-acetylglucosamine-modified proteins. Journal of Biotechnology, 2013, 168(4), 315-323.
-
K. J. Roberson, P. N. Brady, M. M. Sweeney, and M. A. Macnaughtan,* Methods to identify the NMR resonances of the 13C-dimethyl N-terminal amine on reductively methylated proteins. Journal of Visualized Experiments, 2013, 82, e50875. Video.
-
I. Jancan† and M. A. Macnaughtan,* Acid dissociation constants of uridine-5′-diphosphate compounds determined by 31phosphorus nuclear magnetic resonance spectroscopy and internal pH referencing. Analytica Chimica Acta, 2012, 749, 63-69.
-
M. Schimpl, X. W. Zheng, V. S. Borodkin, D. E. Blair, A. T. Ferenbach, A. W. Schüttelkopf, I. Navratilova, T. Aristotelous, O. Albarbarawi, D. A. Robinson, M. A. Macnaughtan, and D. M. F. van Aalten,* O-GlcNAc transferase invokes nucleotide sugar pyrophosphate participation in catalysis. Nature Chemical Biology, 2012, 8(12), 969-974. PDB ID: 4AY5 "Human O-GlcNAc transferase (OGT) in complex with UDP and glycopeptide", PDB ID: 4AY6 "Human O-GlcNAc transferase (OGT) in complex with UDP-5SGlcNAc and substrate peptide"
-
H. Takeuchi, R. C. Fernández-Valdivia, D. S. Caswell, A. Nita-Lazar, S. Kakuda, N. A. Rana, T. Garner, T. Weldeghiorghis, M. A. Macnaughtan, H. Jafar-Nejad, and R. S. Haltiwanger,* Rumi functions as both a protein O-glucosyltransferase and a protein O-xylosyltransferase. Proceedings of the National Academy of Sciences, 2011, 108(40), 16600-16605.
-
H.-W. Lee, G. Wylie, S. Bansal, X. Wang, A. W. Barb, M. A. Macnaughtan, A. Ertekin, G. T. Montelione, and J. H. Prestegard,* Three-dimensional structure of the weakly associated protein homodimer SeR13 using RDCs and paramagnetic surface mapping. Protein Science, 2010, 19(9), 1673-1685.
-
S. Zheng, G. Kaur, H. Wang, M. Li, M. Macnaughtan, X. Yang, S. Reid, J. Prestegard, B. Wang, and H. Ke,* Design, synthesis, and structure-activity relationship, molecular modeling and NMR studies of a series of phenylalkyl ketones as highly potent and selective phosphodiesterase-4 inhibitors. Journal of Medicinal Chemistry, 2008, 51, 7673-7688.
-
M. A. Macnaughtan, F. Tian, S. Liu, L. Meng, S. Park, P. Azadi, K. W. Moremen, and J. H. Prestegard,* 13C-Sialic acid labeling of glycans on glycoproteins using ST6Gal-I. Journal of the American Chemical Society, 2008, 130, 11864-11865.
-
M. A. Macnaughtan, T. Weldeghiorghis, X. Wang, S. Bansal, F. Tian, D. Wang, H. Janjua, K. Cunningham, L.-C. Ma, R. Xiao, J. Liu, M. C. Baran, G. V. T. Swapna, T. B. Acton, B. Rost, G. T. Montelione, and J. H. Prestegard, Solution NMR structure of the homodimer protein YVFG fromBacillus subtilis, Northeast Structural Genomics Consortium Target SR478. Protein Data Bank, 2007, PDB ID: 2JS1.
-
M. A. Macnaughtan, M. Kamar, G. Alvarez-Manilla, A. Venot, J. Glushka, J. M. Pierce, and J. H. Prestegard,* NMR structural characterization of substrates bound to N-acetylglucosaminyltransferase V. Journal of Molecular Biology, 2006, 366, 1266-1281.
-
M. A. Macnaughtan, A. Kane,† and J. H. Prestegard,* Mass spectrometry assisted assignment of NMR resonances in 13C-reductively methylated proteins. Journal of the American Chemical Society, 2005, 127, 17626-17627.
-
M. A. Macnaughtan, A. P. Smith, P. B. Goldsbrough, R. E. Santini, and D. Raftery,* NMR difference spectroscopy with a dual saddle-coil difference probe. Analytical and Bioanalytical Chemistry, 2004, 378, 1520-1527.
-
M. A. Macnaughtan, T. Hou, J. Xu, and D. Raftery,* High-throughput nuclear magnetic resonance analysis using a multiple coil flow probe. Analytical Chemistry, 2003, 75(19), 5116-5123.
-
M. A. Macnaughtan, T. Hou, E. MacNamara, R. E. Santini, and D. Raftery,* NMR difference probe: A dual-coil probe for NMR difference spectroscopy. Journal of Magnetic Resonance, 2002, 156 (1), 97-103.
-
A. R. Pradhan, M. A. Macnaughtan, D. Raftery, Preparation of zeolites supported on optical microfibers. Chemistry of Materials, 2002, 14(7), 3022-3027.
-
T. Hou, J. Smith, E. MacNamara, M. Macnaughtan, and D. Raftery,* Analysis of multiple samples using multiplex sample NMR: Selective excitation and chemical shift imaging approaches. Analytical Chemistry, 2001, 73 (11), 2541-2546.
-
D. Raftery,* S. Pilkenton, C. V. Rice, A. Pradhan, M. Macnaughtan, S. Klosek and T. Hou, Investigation of environmental photocatalysis by solid-state NMR spectroscopy, in 12th International Congress on Catalysis, 2000, 130A, 671-676.
-
A. R. Pradhan, M. A. Macnaughtan, and D. Raftery,* Zeolite-coated optical microfibers for intrazeolite photocatalysis studied by in situ solid-state NMR. Journal of the American Chemical Society, 2000, 122, 404-405.