Model Assessment
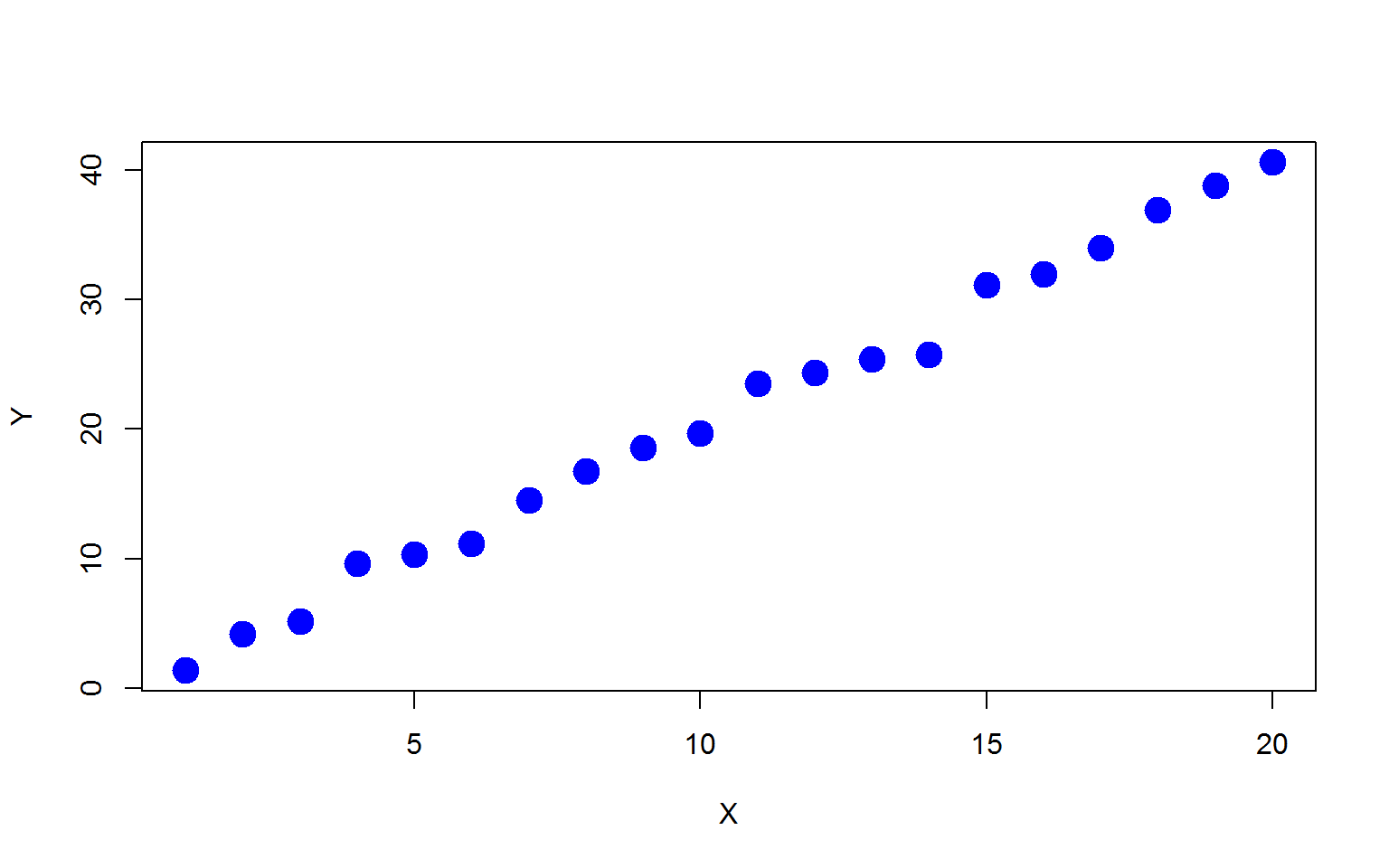
First, let’s generate a simple simulation example with \(x=1,2,\cdots,20\) and \(y\) is a linear function of \(x\) with some additive random noise:
\[ y_i=2x_i+\epsilon_i, \quad \epsilon \sim N(0,1). \]
n=20
x=1:n
set.seed(1)
e=rnorm(n,mean=0,sd=1)
y=x*2+e
plot(x,y,xlab="X",ylab="Y",pch=19,col="blue",cex=2)
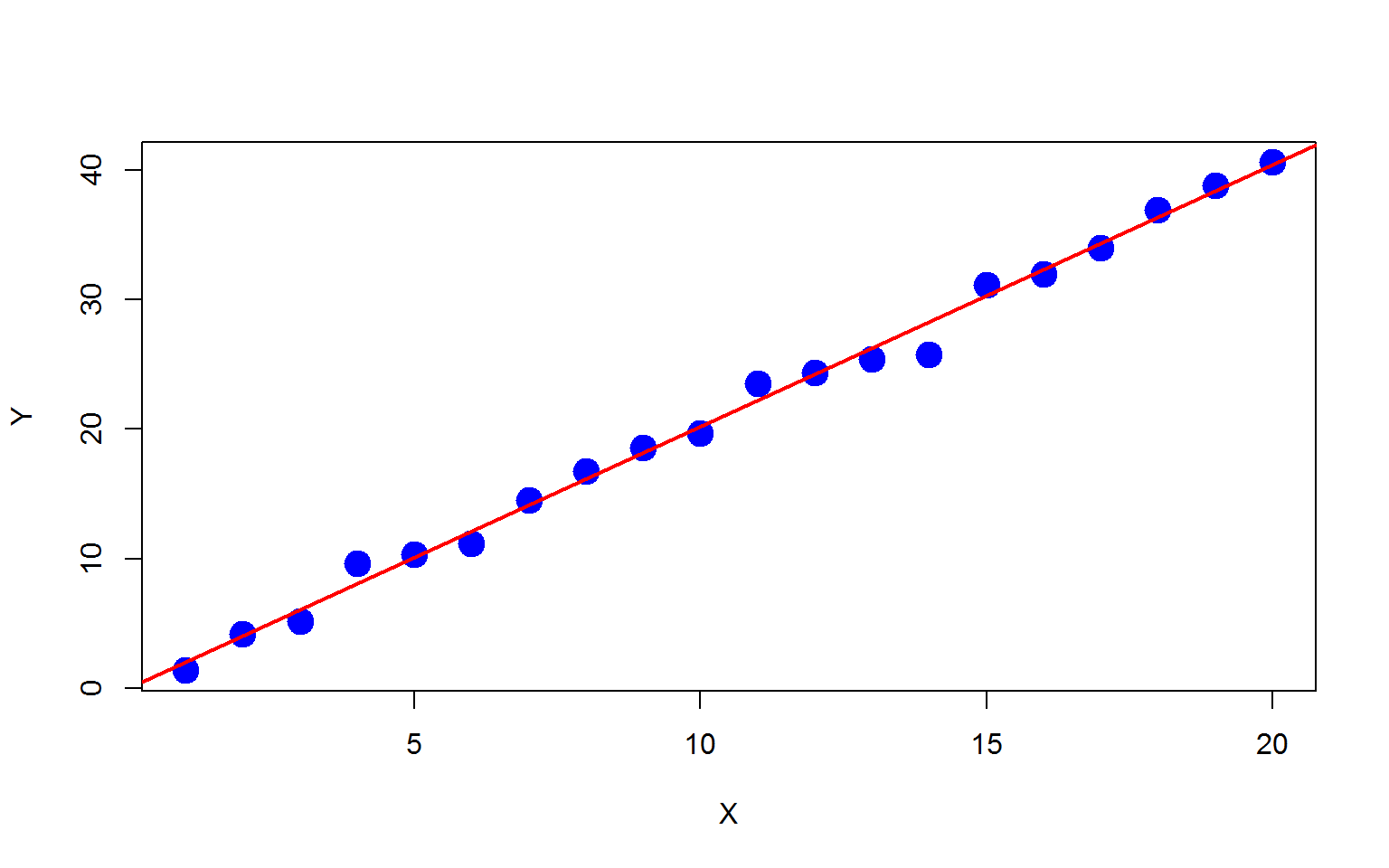
Then we fit a simple linear regression on the dataset.
f=lm(y~x)
plot(x,y,xlab="X",ylab="Y",pch=19,col="blue",cex=2)
abline(f,col="red",lwd=2)
f$fit
## 1 2 3 4 5 6 7
## 1.985490 4.007072 6.028655 8.050237 10.071820 12.093402 14.114985
## 8 9 10 11 12 13 14
## 16.136568 18.158150 20.179733 22.201315 24.222898 26.244480 28.266063
## 15 16 17 18 19 20
## 30.287645 32.309228 34.330810 36.352393 38.373976 40.395558
f$residual
## 1 2 3 4 5 6
## -0.6119435 0.1765711 -0.8642834 1.5450435 0.2576879 -0.9138708
## 7 8 9 10 11 12
## 0.3724441 0.6017572 0.4176313 -0.4851210 1.3104660 0.1669455
## 13 14 15 16 17 18
## -0.8657208 -2.4807627 0.8372856 -0.3541615 -0.3470007 0.5914432
## 19 20
## 0.4472457 0.1983433
mean(f$residual^2) #MSE on training set
## [1] 0.7768427
In-sample Optimism
The MSE on the training set is obviously smaller than the true value (since this is a simulation example, we do know the MSE.
set.seed(2)
e2=rnorm(n,mean=0,sd=1)
y2=x*2+e2
mean((y2-f$fit)^2) #MSE on new responses
## [1] 1.061651
You can see that the MSE on the training set is too optimistic. Now let’s find out the expected loss on the training samples for new response values \[ \frac{1}{n}\sum_{i=1}^n E_{Y^0}\left [ L(Y_i^0, \hat{f}(x_i))|\mathcal{T}\right ]. \]
iter=1000
mse=rep(0,iter)
for (i in 1:iter){
set.seed(i+1)
e3=rnorm(n,mean=0,sd=1)
y3=x*2+e3
mse[i]=mean((y3-f$fit)^2)
}
mean(mse) #Estimate of the expected loss on training set with new Ys
## [1] 1.043732
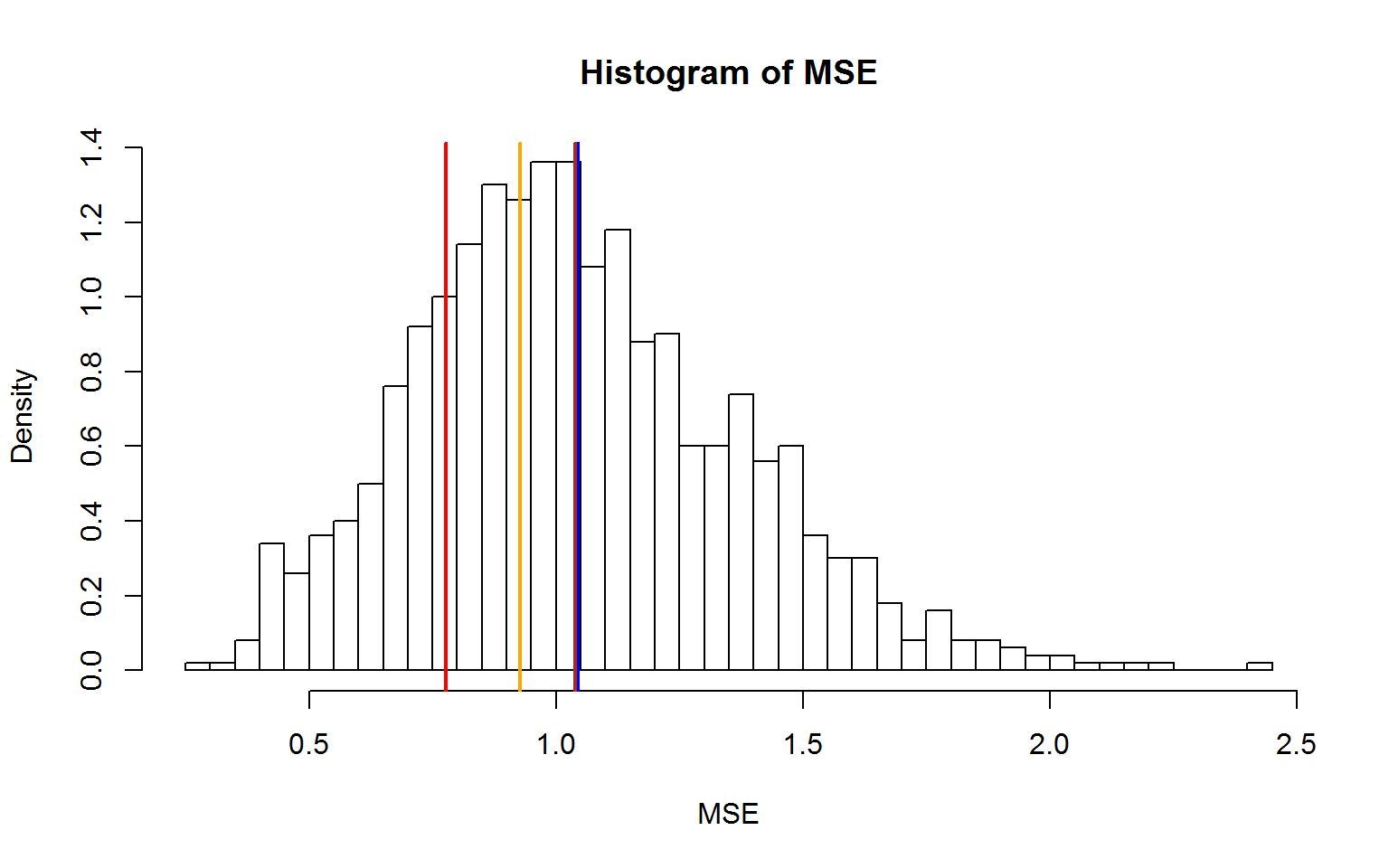
hist(mse,nclass=50,xlab="MSE",ylab="Density",freq=F,main="Histogram of MSE")
abline(v=mean(f$residual^2),col="red",lwd=2)
abline(v=mean(mse),col="blue",lwd=2)
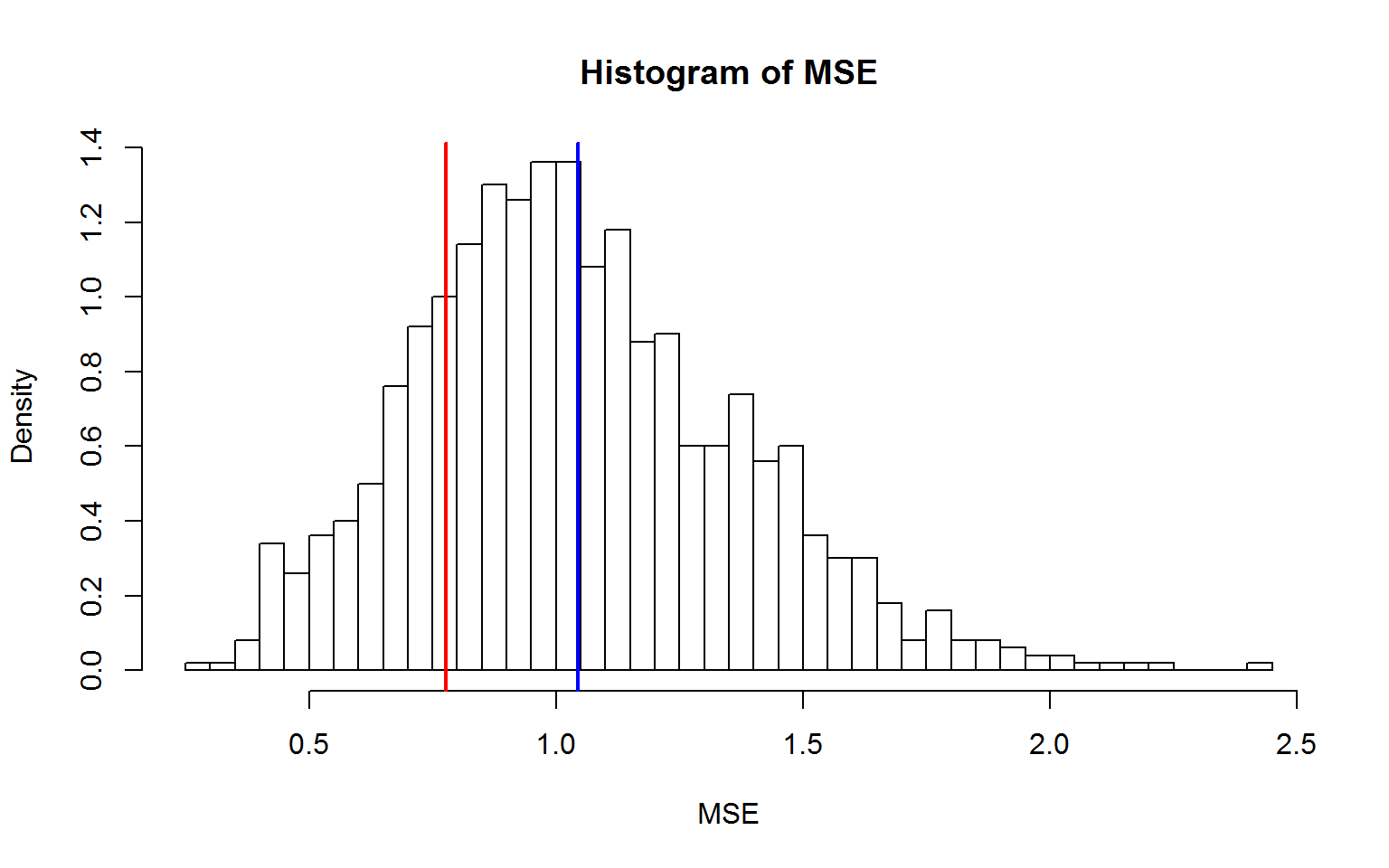 In-sample optimism is defined as the difference between the expected loss on the training
samples for new response values and the training error
In-sample optimism is defined as the difference between the expected loss on the training
samples for new response values and the training error
\[
op=\frac{1}{n}\sum_{i=1}^n E_{Y^0}\left [ L(Y_i^0, \hat{f}(x_i))|\mathcal{T}\right
] - \overline{err}.
\]
If we can find out the in-sample optimism, we can have a better estimate of the expected loss on the training samples for new response values.
\[
\frac{1}{n}\sum_{i=1}^n E_{Y^0}\left [ L(Y_i^0, \hat{f}(x_i))|\mathcal{T}\right ]=op+\overline{err}.
\]
#In-sample optimism
op=mean(mse)-mean(f$residual^2)
op
## [1] 0.2668894'
#A better estimate of performance on test x values
op+mean(f$residual^2)
## [1] 1.043732
Mallow Cp and AIC are developed based on the above idea with the fact that
\[
E_y(op)=\frac{2}{n} \sum_{i=1}^n Cov(\hat{y},y_i).
\]
K-fold Cross-validation
We can estimate the generalization performance through cross-validation. For linear models, we can easily calculate the leave-one-out CV without fitting models n times.
\[
\frac{1}{n}\sum_{i=1}^n [y_i-\hat{f}^{-i}(x_i)]^2 = \frac{1}{n}\sum_{i=1}^n \left[
\frac{y_i-\hat{f}(x_i)}{1-H_{ii}}\right ]^2, \quad where \quad \hat{f}=Hy.
\]
#Hat-diagonal values (leverage)
hat.diag=influence(f)$hat
#LOOCV: PRESS (predicted residual error sum of squares)
press=mean(((y-f$fit)/(1-hat.diag))^2)
press
## [1] 0.927807
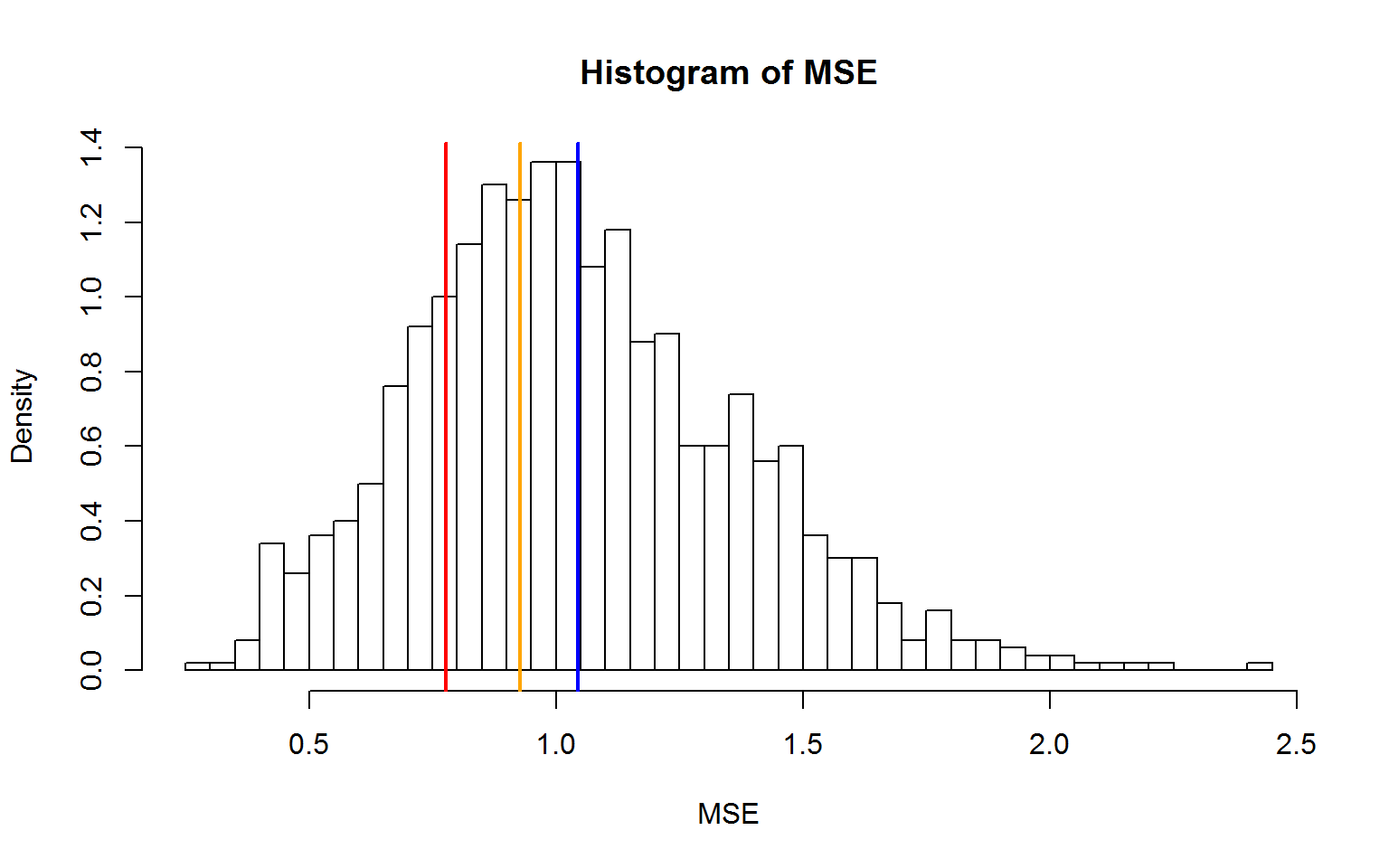
hist(mse,nclass=50,xlab="MSE",ylab="Density",freq=F,main="Histogram of MSE")
abline(v=mean(f$residual^2),col="red",lwd=2)
abline(v=mean(mse),col="blue",lwd=2)
abline(v=press,col="orange",lwd=2)
Out-of-bag (OOB) Samples in Bootstrapping
we can also use out-of-bag (OOB) samples in bootstrapping to estimate the generalization performance of a model. Let’s first see what the OOB samples are.
#Use Out-of-bag (OOB) estimate to evaluate performance
set.seed(1)
boot.id=sample(1:20,size=20,replace=T)
boot.id
## [1] 6 8 12 19 5 18 19 14 13 2 5 4 14 8 16 10 15 20 8 16
setdiff(1:20,boot.id) #out-of-bag samples
## [1] 1 3 7 9 11 17
Let’s estimate the genalization performance of a model using OOB samples.
iter=1000
mse.boot=rep(0,iter)
for (i in 1:iter){
set.seed(i+1)
boot.id=sample(1:20,size=20,replace=T)
f2=lm(y[boot.id]~x[boot.id])
oob.id=setdiff(1:20,boot.id)
y.hat=x[oob.id]*f2$coef[2]+f2$coef[1]
mse.boot[i]=mean((y[oob.id]-y.hat)^2)
}
mean(mse.boot)
## [1] 1.039492
hist(mse,nclass=50,xlab="MSE",ylab="Density",freq=F,main="Histogram of MSE")
abline(v=mean(f$residual^2),col="red",lwd=2)
abline(v=mean(mse),col="blue",lwd=2)
abline(v=press,col="orange",lwd=2)
abline(v=mean(mse.boot),col="brown",lwd=2)